

- Home
- Companies
- Cambridge Protein Arrays Ltd. (CPAP)
- Products
- HuProt - Human Proteome Microarrays

HuProt - Human Proteome Microarrays
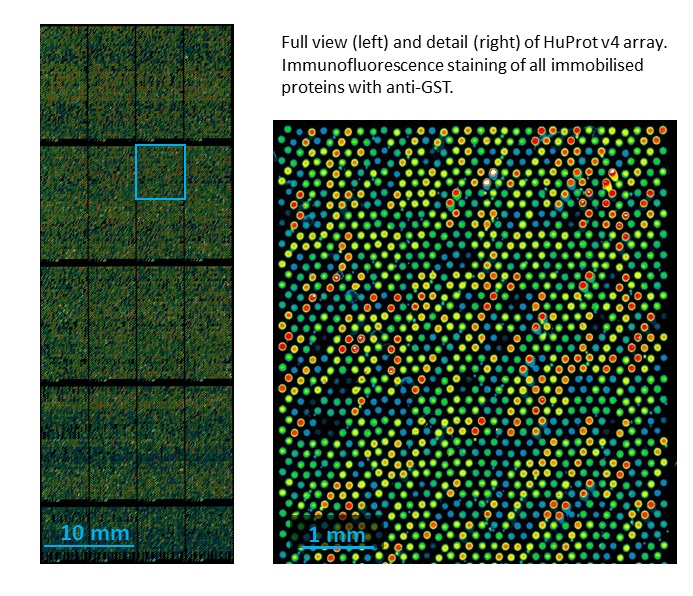
Use the most extensive arrays of full-length human proteins, covering 80% of the human proteome. We are European suppliers for the distribution and service of HuProt Human Proteome microarrays. The new HuProt Human Proteome Microarray v4 offers the largest number of human proteins currently available on a single microarray slide, the majority full length. HuProt arrays contain over 20,000 individually printed proteins, representing more than 16,000 human genes.
The arrays allow screening applications covering more than 80% of the human proteome. Major protein classes and functional contexts of high interest are covered in depth.
Proteins are expressed recombinantly in yeast (S. cerevisiae) as GST fusions, then purified and spotted in duplicate. The complete collection has been resequenced.
Peer reviewed publications using HuProt Human Proteome Arrays in a variety of applications demonstrate protein functionality across the spectrum of immobilised proteins.
We offer HuProt arrays for sale, allowing you to conduct your own experiments and analysis. Alternatively, you may wish to send us your samples and take advantage of our specialist equipment and experience in array handling, assay setup and data analysis, in our HuProt specificity screening service.
Protein content:
- > 20,000 human proteins, representing > 16,000 human protein coding genes (HuProt v4.0)
- Most proteins expressed as full length, several with isoforms.
- Sequences available in dedicated database
- In-depth coverage of major protein classes
- Expressed in Saccharomyces cerevisiae with N-terminal GST-RGS-His6 tag
- GST-based purification
- Spotted assay controls: GST dilution series, biotinylated BSA, mouse IgG anti-biotin, rabbit IgG anti-biotin, mouse IgM, BSA, buffer
- Fluorescent control spots for grid alignment
Array layout:
- Each protein spotted in adjacent duplicates, ≈100 micrometres spot diameter, vertically staggered hexagonal grid
- Printed area: 21.5 mm x 56.5 mm, 4 x 5 subarrays
- Margins (from edge of slide to printed area): top 4 mm, right and left 1.75 mm
- .gal file with spot locations and identities provided with each array batch
- Customised inclusion of additional proteins on request
Slides:
- Standard surface: PATH (Grace Bio-Labs)
- The slides have a nitrocellulose film (Slide dimensions 25 mm x 76 mm x 1 mm
- Barcoded for unambiguous and automated tracking
- Different printing surfaces available on request
Shipping and Storage:
- Shipped on dry ice
- Store at -80°C for 12 months shelf life
Required specialist equipment:
Microarray scanner compatible with 25 mm x 76 mm x 1 mm slides and reflective surfaces (for PATH slides), recommended scan resolution 10 µm per pixel.
Microarray analysis software with capability for handling hexagonal spot grids.
We also provide full service with HuProt arrays: send us your samples, benefit from our expertise and equipment, receive your results. Alternatively, we can scan and analyse HuProt arrays probed by you.
Proteome-wide coverage across relevant protein classes.
HuProt v4 arrays offer an unrivalled coverage of 80% of the human proteome, on a single slide for probing in one experiment.
Overview of coverage of protein classes of special interest
Coverage analysis of the proteome and individual protein classes has been performed according to the full proteome list and annotation in The Human Protein Atlas. As members of the classes overlap, the figures in the pie charts are not additive.
Human Proteome Microarrays.
Protein content:
- > 20,000 human proteins, representing > 16,000 human protein coding genes (HuProt v4.0)
- Most proteins expressed as full length, several with isoforms.
- Sequences available in dedicated database
- In-depth coverage of major protein classes
- Expressed in Saccharomyces cerevisiae with N-terminal GST-RGS-His6 tag
- GST-based purification
- Spotted assay controls: GST dilution series, biotinylated BSA, mouse IgG anti-biotin, rabbit IgG anti-biotin, mouse IgM, BSA, buffer
- Fluorescent control spots for grid alignment
Array layout:
- Each protein spotted in adjacent duplicates, ≈100 micrometres spot diameter, vertically staggered hexagonal grid
- Printed area: 21.5 mm x 56.5 mm, 4 x 5 subarrays
- Margins (from edge of slide to printed area): top 4 mm, right and left 1.75 mm
- .gal file with spot locations and identities provided with each array batch
- Customised inclusion of additional proteins on request
Slides:
- Standard surface: PATH (Grace Bio-Labs)
- The slides have a nitrocellulose film (
- Slide dimensions 25 mm x 76 mm x 1 mm
- Barcoded for unambiguous and automated tracking
- Different printing surfaces available on request
Shipping and Storage:
- Shipped on dry ice
- Store at -80°C for 12 months shelf life
Required specialist equipment:
Microarray scanner compatible with 25 mm x 76 mm x 1 mm slides and reflective surfaces (for PATH slides), recommended scan resolution 10 µm per pixel.
Microarray analysis software with capability for handling hexagonal spot grids.
We also provide full service with HuProt arrays: send us your samples, benefit from our expertise and equipment, receive your results. Alternatively, we can scan and analyse HuProt arrays probed by you.
Please get in touch to discuss the best options.
