
Karyosoft Inc.
- Home
- Companies
- Karyosoft Inc.
- Software
- Karyosoft - Version Variant Mining ...

Karyosoft - Model Variant Mining Studio - Software for SNP and Indel Mining Made Easy and Secure
FromKaryosoft Inc.
A central repository for all your millions of SNPs and Indels. Data organized in project specific and/or organism specific (microbes, animals, plants, humans and more). Hosted on your internal servers or any cloud servers. No more data loss; No need to open on excel. Web-based and user-friendly for anywhere and anytime on any device.
Most popular related searches

- Create the "Project" and "Organism" and upload the vcf files of SNPs and Indels and keep all your millions of SNPs and Indels in an organized way in one-place.
- Serves as a central repository for all your SNPs and Indels and easy retrieval of mutation information quickly at anytime and anywhere to drive innovations.

- Select the "Project" and the "Organism" from which the SNP and Indel data were generated
- Either by clicking "Browse" or "Drag the files" and choose vcf files of SNPs and Indels.
- Click "Save" for uploading the data.
- SNPs and Indels from the vcf files will automatically be uploaded into "SNPs" and "Indels" tabs.

- Just click "Explore" in the project of interest in the "Projects" tab.
- The project and organism are already pre-selected, and all your SNPs will be displayed on "SNPs" tab.
- Click on the "Indels" tab to view all your Indels.

- Search the millions of SNPs and Indels in "SNPs" and Indels" tab by gene name, annotation effect, putative impact, chromosome number
- With "Advanced Search", you can search by giving genomic location interval (For e.g. 10,000 - 30,000) to retrieve SNPs and Indels in that specific interval.
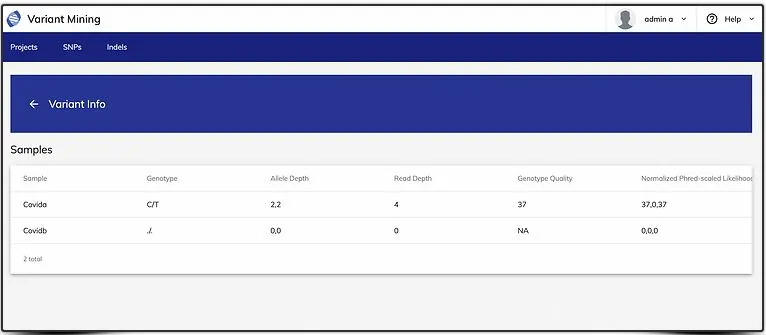
- By clicking the "Chromosome" in the first column, you can find the "Genotype" calls of all your samples in homozygous or heterozygous state including missing data.
- By clicking on the ">", you can find the annotation information of that specific locus.
- Easy download of only the selected SNPs and Indels" at a specific genomic location by clicking "Download".
